Population and hierarchical genetic structure of Badis badis (Hamilton-Buchanan, 1822) in sub-Himalayan Terai region of West Bengal, India 
 Author
Author  Correspondence author
Correspondence author
International Journal of Aquaculture, 2015, Vol. 5, No. 27 doi: 10.5376/ija.2015.05.0027
Received: 20 Jun., 2015 Accepted: 21 Jul., 2015 Published: 16 Sep., 2015
Mukhopadhyay T. and Bhattacharjee S., 2015, Population and hierarchical genetic structure of Badis badis (Hamilton- Buchanan, 1822) in sub-Himalayan Terai region of West Bengal, India, International Journal of Aquaculture, 5(27): 1-10
Badis badis is a threatened ornamental freshwater fish in the Indian scenario. But the population genetic background of this ichthyofauna is largely unexplored in the eastern sub-Himalayan hotspot region of West Bengal state of India, known as the Terai. We have studied six populations from two major streams (Mahananda and Balason) of the region through RAPD fingerprinting. The allelic richness, Shannon’s Information index and a measure of evenness were calculated for each population. The SHE analysis revealed the change in diversity pattern in a spatial scale. Our results indicated that the allelic richness decreased and the evenness increased in the populations as the streams drained from higher to a lower altitude. The Nei’s genetic distance, genetic identity and UPGMA dendrogram revealed that the six populations formed two distinct groups. The principal component analysis also supported the UPGMA dendrogram. The pair-wise gene differentiation and gene flow between the populations showed the hierarchical genetic structure of Badis badis in the studied region.
Introduction
Badis badis (Hamilton-Buchanan 1822) (Actinopterygii, Perciformes, Badidae) a tropical, benthopelagic, ornamental freshwater fish has recently been included within the vulnerable category in the list of threatened freshwater fishes of India by National Bureau of Fish Genetic Resources (NBFGR, Lucknow, India) (Lakra et al, 2010). However, there have been insufficient studies with regard to the spatial genetic architecture of this fish species, especially in the eastern sub-Himalayan hotspot region of West Bengal, India, known as the Terai.
Genetic variability or diversity is an essential for the fitness as well as survival of the whole population. Therefore the loss of genetic diversity of a species reduces its capability for adaptation and increases the risk of extinction (Frankham, 1995; Caughley and Gunn, 1996; Avise and Hamrick, 1996; Landweber and Dobson, 1999). The endangered/vulnerable organisms having small population size may experience a continuous reduction in the level of genetic variation. However, maintaining genetic variation in endangered species is essential to ensure its adaptation, expansion and proper reestablishment in natural conditions, if required. The study of genetic variability within and between local populations is extremely useful to gain information on the individual identity, breeding patterns, the degree of relatedness and genetic variability within as well as between them. Genetic variations can be assessed by means of DNA polymorphisms. RAPD fingerprinting has been used to evaluate genetic diversity and also in subspecies identification (Wasko et al., 2002; Leuzzi et al., 2004; Hatanaka and Galetti, 2003; Feng et al., 2007; Islam and Alam, 2004; Garg et al., 2009; Garg et al., 2010; Alam et al., 2010; Affonso and Galetti, 2007; Amavet et al., 2009; Prior et al., 1997; Brahmane et al., 2006; Bardakci and Skibinski, 1994; Brahmane et al., 2008; Yamazak et al., 2005). A very limited genomic research has been carried out in Badis badis species till date and insufficient genomic information is available to perform other sophisticated fingerprinting techniques where whole genomic sequence is necessary; therefore, RAPD fingerprinting was the best alternative for genetic diversity analyses. A number of diversity indices have been widely used by different investigators to quantitatively express diversity. Shannon’s index of diversity (H´ or I) can be decomposed into two functional components viz.; species richness (E) and evenness (S). These three components add up to SHE analysis that may enable the delineation of change in the diversity pattern (Buzas and Hayek, 1996). This method allows researchers to examine the evenness component separately from the richness and vice-versa in a single step process (Buzas and Hayek, 1998).
To answer the questions regarding the genetic variation in any subdivided population or in the metapopulation, it is very essential to focus our attention towards the “evolutionary functional unit” and a population seems the most reasonable level at which genetic conservation intervention should take place. The population is where the local adaptation and genetic changes occur over generations, therefore allopatric/sympatric population is of great interest. In a noteworthy book on conservation biology, Meffe and Carroll in 1997 pointed out one approach to define ‘evolutionary functional unit’ in a genetic perspective through a hierarchical genetic analysis of subdivided populations.
We have previously investigated the present status of the available genetic diversity in the Badis badis populations in the Terai region of West Bengal, India (Mukhopadhyay and Bhattacharjee, 2014a). We have also reported the genetic diversity within and between the populations of Badis badis and also determined the total available genetic diversity present in the Mahananda-Balason river system of the Terai region of sub-Himalayan West Bengal, India. Based on this firsthand information we have carried out further analyses to extract as much as possible genetic information regarding this threatened ichthyofauna. The objectives of the present study was to (1) ascertain the genetic distance and genetic relatedness of the different populations of Badis badis from the major river streams of the Terai region of sub-Himalayan West Bengal, India, (2) determine the changes in the diversity pattern through SHE analysis among different populations of Badis badis from the streams of the region, and (3) ascertain hierarchical genetic structure among different Badis populations.
1 Results
1.1 RAPD Profile
Twenty-two RAPD primers generated in total 199 amplified fragments from thirty individuals, corresponding to 6 separate riverine collection sites (Table 1). The number of amplified fragments ranged from 6 (OPA-09, OPA-13, OPB-11 and OPB-18) to 13 (OPA-16). Considering all the primers, the size ranges of the amplified fragments ranged from 150 to 2200 base pairs (Table 1).
1.2 Within and between- population diversity and genetic differentiation
The allelic richness or observed number of alleles ranged from 1.2915± 0.4556 in TR-2 to 1.3769 ± 0.4858 in TR-3 populations. While Shannon’s Information index (H´ or I) was highest (0.2205 ± 0.2950) in the TR-3 population (Table 2) and the TR-2 population showed its lowest value (0.1648 ± 0.2691) (Table 2). The highest value of evenness (0.922561) was found in the TR-1 population and the lowest value of evenness (0.904002) was found in the TR-6 population (Table 2).
|
|
|
|
The expected (HT) and mean heterozygosity (HS); diversity among populations (DST); degree of gene differentiation (FST); and estimates of gene flow (Nm) were estimated in pairwise populations. The expected heterozygosity and mean heterozygosity ranged from 0.1764 ± 0.0387 to 0.3124 ± 0.0362 and from 0.1183 ± 0.0178 to 0.1416 ± 0.0229 respectively (Supplementary Table 1). The highest observed value of inter-population diversity (0.1848) was between the populations of TR-1 and TR-5 and the lowest value (0.0452) was observed between the populations of TR-4 and TR-5 (Supplementary Table 1). The degree of gene differentiation was highest (0.5996) between the populations of TR-1 and TR-2 whereas the degree of gene differentiation between the populations of TR-4 and TR-5 was lowest (0.2560) (Supplementary Table 1 and Figure 2). Moreover the populations of TR-4 and TR-5 showed maximum level of gene flow (1.4534) between them while the extent of gene flow was minimum between the populations of TR-1 and TR-2 (0.3339) (Supplementary Table 1 and Figure. 2). When total populations were considered the total genetic diversity (HT) was 0.2983 ± 0.0203 and the total genetic differentiation coefficient (FST) among the populations was 0.5629 (Supplementary Table 1).
1.3 Genetic distance and relatedness
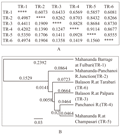
The Nei’s genetic distance between the populations of TR-1 and TR-5 was maximum (0.5350) and the distance between the populations of TR-4 and TR-5 was minimum (0.0928) (Figure 3A). The genetic identity was highest (0.9114) between the TR-4 and TR-5 populations but the identity between the TR-1 and TR- 5 populations was lowest (0.5857) (Figure 3A). Thus it was observed from the Nei’s genetic distance and identity as well as from the UPGMA dendrogram that the TR-1 and TR-5 populations were more distant from each other while the TR-4 and TR-5 populations were more closely related to each other (Figure 3B). The dendrogram indicated that the six populations formed two groups, one group consisting five populations viz., TR-2, TR-3, TR-4, TR-5 and TR-6 whereas the population of TR-1 formed a single outgroup (Figure 3B). The two-dimensional PCoA plot (Figure 4) gave a clearer picture of clustering of 6 Badis badis populations in this region. The four populations viz., TR-3; TR-4; TR-5; TR-6 formed a cluster whereas TR-1 and TR-2 constituted two separate groups (Figure 4). Therefore, the PCoA analysis was largely in agreement and confirmation with the UPGMA dendrogram.
|
|
1.4 SHE analysis
SHE analysis revealed a clear distribution of three biodiversity components richness (S), diversity (H´) and evenness (E) of the Badis badis gene pool into six different riverine populations of the Terai regions. We found that the lnS and H´ were highest in TR-3 (0.319835 and 0.2205) and lowest in TR-2 population (0.255804 and 0.1648 respectively) (Figure 5 lower panel). The lnE value was highest in TR-1 population (-0.8060) and lowest in TR-6 population (0.10092) (Figure 5 lower panel). The SHE analysis plot revealed the observed pattern for distribution of three components viz., S (richness), H´ (Shannon's Information index) and E (evenness) in relation to six different populations. We divided the six riverine populations in three groups [TR-1, TR-3, TR-6 constituting first group (Plot A); TR-1, TR-2, TR-5 constituting second group (Plot B); TR-1, TR-2, TR-4 constituting third group (Plot C)] based on the continuity of the water flow through the river stream (Figure 5). Since water flows from higher to lower altitude there is an expected trend of increase in S (richness) and H´ (Shannon's Information index) but decrease in E (evenness) in all the three analyses as one goes upstream (Figure 5 upper panel).
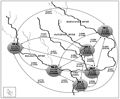 Figure 6 Genetic hierarchical model of six different populations of Badis badis. The dotted arrows indicate the gene differentiation (FST) and gene flow (Nm) (within parentheses) (see inner box). The shaded circles indicate the collection sites; S= richness; H= Shannon Information index (H´); E= measure of evenness. Major streams are marked in dark lines and minor streams are indicated in faded lines |
2 Discussions
RAPD technique has been widely used to ascertain the available gene pool of different subdivided populations of a species that may have arisen either by means of selection pressure or as a result of genetic drift (Fuchs et al., 1998). The ornamental fish Badis badis has been considered to be a vulnerable fish species in the Indian scenario demanding conservation, management and stock enhancement in the Terai region of West Bengal, India. The region is within a biodiversity hotspot; therefore, acquiring information regarding the population genetic structure of this species might be helpful to the development of suitable conservation strategies. To our knowledge, the present study is the first attempt to explore and also to investigate the present status population- specific genetic relationship of this species, changes in the diversity pattern and also map the genetic hierarchy in this sub-Himalayan hotspot region of northern West Bengal, India.
Since Shannon’s index of diversity can be decomposed into two metric components namely species richness (E) and evenness (S) i.e., (H´= lnE+ lnS) (Buzas and Hayek, 1996) sometimes it is difficult to interpret whether the diversity index is influenced by greater/lower richness or greater/lower evenness values or both. However, this decomposition also allows us to analyze the change in diversity pattern though different subpopulations. We have found that the diversity (H´), richness (S) and evenness (E) have varied across all six populations (Figure 5 and 6) which are most obvious in naturally subdivided populations. Therefore the SHE analysis can deduce the change in the diversity pattern of populations along a gradient and also look for the breaks in the pattern that indicate the change in diversity of the population (Buzas and Hayek, 1998). Hayek and Buzas (1997) pointed out that often the diversity (H´) changes because the differences between richness (S) and evenness (E) do not offset each other (i.e., H´1 ≠ H´2, S1 ≠ S2, E1 ≠ E2 , where 1 and 2 in suffix are any two population) and such SHE plot is log normal one. SHE analysis appears to be a useful approach for defining the diversity; moreover, it allows a high resolution visualization of the changes in diversity in a temporal as well as spatial scale. Our data revealed that as the river streams converged from higher to lower altitude, the diversity and richness of the Badis badis populations decreased and evenness increased (Figure 5, Plots A, B and C). This decrease in diversity and richness within the gene pool of the Badis population may be due to flow pattern disturbances and human interferences (such as fishing and pesticide run-offs from adjacent tea gardens in the hilly areas of lower Himalayas) as the river streams flow from higher to lower altitudes. All of these causes can culminate in to the observed decline and change in diversity pattern and richness in Badis badis populations across the river stream along the altitudinal gradient.
Fixation index or FST is a measure of genetic divergence among subpopulations that ranges from 0 (when all subpopulations have equal allele frequencies) to 1 (when all the subpopulations are fixed for different alleles) (Allendorf et al., 2013). We found that the FST value was highest between the TR-1 and TR-2 population and consequently lowest gene flow existed between them (Supplementary Table 1 and Figure 5, 6). The FST and Nm values between TR-1 and TR-5 population (0.5917 and 0.3450) and that between TR-1 and TR-4 population (0.5466 and 0.4147) indicate that there is reduced gene flow from the Mahananda (TR-2 and TR-5 population) and Panchanoi (TR-4 population) rivers to the Fulbari barrage (TR-1) population (Supplementary Table 1 and Figure 5, 6). However in contrast, higher gene flow is observed between Balason (TR-3 and TR-6) and Fulabari barrage (TR-1) population (Supplementary Table 1 and Figure 5, 6). The differences observed in the FST and Nm values between Mahananda, Panchanoi and Balason river populations may be attributable to several factors, viz, water flow pattern, water volume, fishing extent and other possible anthropogenic and/or geological activities. The FST value was lowest (0.2560) and gene flow (1.4534) was highest between the Panchnoi River (TR-4) and Champasari (TR-5) populations (Supplementary Table 1 and Figure 5, 6). Although not linked at the upstream region there is a possibility that these two regions might be linked during the monsoon season though flood plains causing mixing of individuals between these two regions. This intermixing of individuals as well as inter-breeding may cause lower gene differentiation and high rate of gene flow among these two sites (TR-4 and TR-5). Moreover when we compared the six populations in a pair-wise manner, we found that the TR-1 population lies more divergent from other five populations and gene flow was also lowered between TR-1 with other five populations. The same was also true for TR-2 with other five populations. This confirms an independent and gradual isolated genetic differentiation across the population of Badis badis. The independent genetic differentiation of this population may be attributed to the geographical distance. The Nei’s genetic distance and genetic similarity exposed a distinct spatial relationship between the six Badis badis populations. In the UPGMA based dendrogram TR-1 population formed an outgroup which might be due to the convergence of all other three river streams along with the Badis badis gene pool to the barrage before being released to Bangladesh. In general, this spatial genetic architecture delineates the genetic hierarchical structure of Badis badis populations across the main river streams of the Terai region.
We have involved a population of five individuals each from each collection site primarily because of the dwindling population structure of the species. However to compensate this weak point of the study we have used a set (twenty-two) primers to generate a robust data matrix. Our study revealed a distinct pattern of genetic diversity and genetic divergence that exists across the six Badis badis populations in the study area. The populations were allopatrically distinct or separated from each other although gene flow was still evident in the populations which may prevent the population from inbreeding depression and consequently maintaining their survival. Moreover, it was also found with the help of SHE analysis, FST analysis and dendrogram of the six populations of Badis badis, that a genetic hierarchical structure was present in the populations. SHE analysis was employed to separate species diversity into its richness and evenness components. The SHE analysis revealed that the change in the genetic diversity of the Badis populations was due to altitudinal variation in the genetic richness and evenness in the populations. We have previously reported that the genetic diversity of the Badis badis population in the major stream (Mahananda and Balason river) of Terai region was declining. Several anthropogenic interferences like indiscriminate fishing, effluents from adjacent households and factories, heavy use of fertilizers and pesticides in the nearby tea gardens or excavation of river beds along the banks for sand and gravel all may be responsible for the decline of the overall diversity of this threatened ornamental fish species in the Terai region of West Bengal, India. Therefore, proper steps should be taken to improve the environment to avoid further losses. The genetic data obtained from the present study lend support to the view that there is a scope of stock improvement for this threatened ornamental fish species.
3 Materials and Method
3.1 Survey and Sample Collection
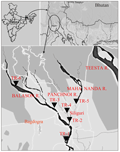
An extensive survey has been carried out in different spots of the major streams (i.e. Mahananda and Balason river) of the Terai Region of West Bengal, India. A total thirty fish (five individuals from each collection spots) tissue samples of Badis badis were collected from six different spots and geographic co-ordinates were recorded with the help of GPS (eTrex Vista HCx, Garmin, USA). We have previously reported that the genetic diversity of this fish population was low (Mukhopadhyay T, Bhattacharjee, 2014a). Therefore, a limited number of individuals were involved for further population genetic analyses primarily because of the dwindling population structure of this species in the study region. The collection spots were as follows: TR-1, TR-2, TR-3, TR-4, TR-5 and TR-6. The local names and geographical co-ordinates of the collection spots are mentioned in Figure 1. Fishes were identified according to Talwar and Jhingran (1991).
3.2 Isolation of high molecular weight DNA and Quantification
Genomic DNA (gDNA) was extracted noninvasively from tiny amount of tissue samples (10-15 mg of fin clips from the caudal and ventral portions and/or 25-30 pieces of scales from the dorsal portion) from the live Badis badis following Mukhopadhyay and Bhattacharjee (2014b). Briefly, tissues were chopped finely and then incubated at 52ºC for 60 min in a lysis buffer containing 15 µl Proteinase K (20 mg/ml) and 30 µl 20% SDS. After the incubation, each lysate was purified by phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v).
3.3 Primer Selection
Forty arbitrary decamer primers of random sequences (Kit-A and Kit-B, twenty primers from each kit) were purchased from Imperial Life Science Pvt. Ltd., India. Firstly, six different populations were screened with the forty primers and finally twenty-two (10 primers from Kit-A and 12 primers from Kit-B) were selected for further analyses on the basis of the variability and reproducibility of the bands obtained (Table 1).
3.4 RAPD-PCR and documentation of amplified products
RAPD analyses were performed in a 96 well Peltier Thermal Cycler (Applied Biosystems 2720, Life Technologies, USA) in a final reaction volume of 25 µl, each containing a final concentrations of ~100-150 ng of isolated gDNA, 1.6 pM of OPA or OPB primer, 1X Standard Taq Polymerase buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2) (NEB, USA), 200 µM of each dNTPs (dATP, dTTP, dCTP, dGTP) (NEB, USA), and one unit of Taq DNA Polymerase (NEB, USA). PCR cycling programs were as follows: initial denaturation at 94º C for 5 min followed by 40 cycles of 94 ºC, 1 min for denaturation; 35 ºC, 1 min for annealing; 72 ºC, 2 min for elongation and finally an extension at 72 ºC for 10 min. The amplified products were electrophoresed in an ethidium bromide (0.5 µg/ml) pre-stained 1.4 % (w/v) agarose gel (Lonza, Basel, Switzerland) at a constant voltage 100 V and current 100 mA in TAE buffer (40 mM Tris-HCl, pH 8.0; 20 mM Acetic acid; 1 mM EDTA, pH 8.0) using BenchTop Labsystems BT-MS-300, Taiwan electrophoretic apparatus. Molecular weight of each band was estimated using a standard 100 base pair ladder (NEB, USA) and/or Lambda EcoRI/HindIII double digest DNA size marker (NEB, USA). The gels were visualized on the UV-transilluminator (Spectroline BI-O-Vision®NY, USA) and photographed using a digital camera.
3.5 RAPD data analyses
A robust RAPD dataset from six Badis badis populations were analyzed for assessing inter-population genetic variability, genetic differentiation, genetic distance and genetic hierarchy between six populations of Badis badis of Terai region. The RAPD fingerprinting profile were computed and analyzed in the form of binary variables (1= band present or 0= band absent) by direct comparison of the amplified pattern. The binary scores obtained from all the twenty-two primers in the RAPD analyses were then pooled for constructing a single data matrix. The RAPD data was analysed using three software viz., Popgene ver. 1.32 (Yeh et al., 1999), TFPGA ver.1.3 (Miller, 1997) and GenAlEx 6.5 (Peakall and Smouse, 2006; Peakall and Smouse, 2012). The data matrix was used to estimate the observed number of alleles or allelic richness [(1/K) ∑ ni, where K= number of loci and ni = the number of alleles detected per locus], Shannon’s Information Index (H´ or I= -∑ pi log2 pi, where H´ or I is diversity and pi is the frequency of a particular RAPD band) (Lewontin, 1972). To calculate the measure of evenness (E) we have used the exponential function of Shannon’s Index i.e, eH´ and subsequently divided it by allelic richness (S) or observed number of alleles (E= eH´/S, where S is the observed number of alleles or allelic richness). The Shannon’s Information index (H´), measure of evenness (E) and observed number of alleles i.e. richness (S) sum up to SHE analysis (H´= lnE+ lnS) (Hayek and Buzas, 1997; Magurran, 2004). To analyze the inter-population genetic differentiation between six different populations of Badis badis, pair-wise FST values were calculated using the formula FST = 1- HS/HT (where HS is the average expected heterozygosity estimated from each subpopulation and HT is the total gene diversity or expected heterozygosity in the total population as estimated from the pooled allele frequencies) (Wright, 1969). The FST is equivalent to the coefficient of gene differentiation GST (GST=DST/HT). The diversity among populations (DST = HT - HS) was also calculated to ascertain the inter-population diversity between different pair-wise populations. The estimated gene flow (Nm) between the pair-wise populations was also calculated by the formula Nm=0.5(1-GST)/GST (McDonald and McDermott, 1993). The binary matrix prepared from all scored fragments were used to generate Nei's unbiased measures of genetic identity and genetic distance matrix (Nei, 1978) using the software Popgene ver. 1.32 and the output data matrix was also verified separately using the software TFPGA ver. 1.3 and Arlequin ver. 3.1 (Excoffier and Schneider, 2005). The Nei’s genetic distance matrix was subjected to unweighted pair-group method using arithmetical averages (UPGMA) to generate a dendrogram through linkage procedure using the software Phylip ver. 3.69 (Felsenstein, 2005) and FigTree ver.1.3.1 (Rambaut, 2010). The Nei’s genetic distance matrix was then used as the basis for ordination by Principal Co-ordinate Analysis (PCoA), which was performed to show the distribution of the genotypes in a scatter plot using GenAlEx ver. 6.5.
Author’s Contribution
TM performed literature search, data acquisition, experimental studies, data analyses, and manuscript preparation. SB designed study, defined intellectual content and performed data analyses, manuscript editing and review.
Acknowledgments
The work was supported by a research grant from University Grants Commission (UGC), India [MRP Sanction No.: 40-289/2011 (SR)] awarded to corresponding author.
References
Affonso P, Galetti Jr. P.M., 2007, Genetic diversity of three ornamental reef fishes (Families Pomacanthidae and Chaetodontidae) from the Brazilian coast, Brazilian Journal of Biology, 67(4): 925-933
http://dx.doi.org/10.1590/S1519-69842007000500017
Alam M.S., Islam M.S. and Alam M.S., 2010, DNA Fingerprinting of the freshwater Mud Eel, Monopterus cuchia (Hamilton) by Randomly Amplified Polymorphic DNA (RAPD) Marker, International Journal of Biotechnology and Biochemistry, 6(2): 271-278
Allendorf F.W., Luikart G., Aitken S.N., 2013, Conservation and the Genetics of Populations, A John Wiley & Sons, Ltd., Publication, UK
Amavet P., Vilardi J.C., Rosso E. and Saidman B., 2009, Genetic and Morphometric Variability in Caiman latirostris (Broad-Snouted Caiman), Reptilia, Alligatoridae, Journal of Experimental Zoology, 311 A : 258-269
Avise J.C. and Hamrick J.L., 1996, Conservation and Genetics: Case Histories from Nature, Chapman & Hall, New York
http://dx.doi.org/10.1007/978-1-4757-2504-9
Bardakci F. and Skibinski D.O.F., 1994, Application of the RAPD technique in tilapia fish sps and subspecies identification, Heredity, 73:117-123
http://dx.doi.org/10.1038/hdy.1994.110
Brahmane M.P., Mitra K. and Misra S.S., 2008, RAPD fingerprinting of the ornamental fish Badis badis (Hamilton 1822) and Dario dario (Kullander and Britz 2002) (Perciformes, Badidae) from West Bengal, India,. Genetics and Molecular Biology, 31:789-792
http://dx.doi.org/10.1590/S1415-47572008000400028
Brahmane M.P., Das M.K., Sinha M.R., Sugunan V.V., Mukherjee A., Singh S.N., Prakash S., Maurye P. and Hajra A., 2006, Use of RAPD fingerprinting for delineating populations of hilsa shad Tenualosa ilisha (Hamilton, 1822), Genetics and Molecular Research, 5(4):643-652
Buzas M.A. and Hayek L.A.C. 1996, Biodiversity resolution: an integrated approach, Biodiversity Letters 3:40-43
http://dx.doi.org/10.2307/2999767
Buzas M.A. and Hayek L.A.C., 1998, SHE analysis for biofacies identification, Journal of Foriminiferal Research, 28:233-239
Caughley G. and Gunn A., 1996, Conservation Biology in Theory and Practice. Blackwell Science, Cambridge
Excoffier L.G.L. and Schneider S., 2005, Arlequin ver. 3.0: An integrated software package for population genetics data analysis, Evolutionary Bioinformatics Online, 1:47-50
Felsenstein J., 2005, PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Feng Y., Peijun Z., Keeling Y. and Jianhai X., 2007, Genetic variation of natural and cultured stocks of Paralichthys olivaceus by allozyme and RAPD, Chinese Journal of Oceanology and Limnology, 25:78-84
http://dx.doi.org/10.1007/s00343-007-0078-9
Frankham R., 1995, Conservation genetics, Annual Review of Genetics, 29: 305-327
http://dx.doi.org/10.1146/annurev.ge.29.120195.001513
Fuchs H., Gross R., Stein H. and Rottmann O.,1998, Application of molecular genetic markers for the differentiation of bream (Abramis brama L.) populations from the rivers Main and Danube, Journal of Applied Ichthyology, 14:49-55
http://dx.doi.org/10.1111/j.1439-0426.1998.tb00613.x
Garg R.K., Sairkar P., Silawat N., Batav N. and Mehrotra N.N., 2010, Assessment of genetic diversity of Clarias batrachus using RAPD markers in three water bodies of Bhopal, Journal of Environmental Biology, 31: 749-753
Garg R.K., Silawat N., Sairkar P., Vijay N. and Mehrotra N.N., 2009, RAPD analysis for genetic diversity of two populations of Mystus vittatus (Bloch) of Madhya Pradesh, India, African Journal of Biotechnology, 8: 4032-4038
Hatanaka T. and Galetti Jr. P.M., 2003, RAPD markers indicate the occurrence of structured populations in a migratory freshwater fish species, Genetics and Molecular Biology, 26: 19-25
http://dx.doi.org/10.1590/S1415-47572003000100004
Hayek L.A.C. and Buzas M.A., 1997, Surveying natural populations. Columbia University Press, New York
Islam M.S. and Alam M.S., 2004, Randomly amplified polymorphic DNA analysis of four different populations of the Indian major carp, Labeo rohita (Hamilton), Journal of Applied Ichthyology, 20: 407-412
http://dx.doi.org/10.1111/j.1439-0426.2004.00588.x
Lakra W.S., Sarkar U.K., Gopalakrishnan A. and Kathirvelpandian A., 2010, Threatened freshwater fishes of India. Published by National Bureau of Fish Genetic Resources (NBFGR), Lucknow, India
Landweber L.F. and Dobson A.P., 1999, Genetics and the Extinction of Species: DNA and the Conservation of Biodiversity. Princeton University Press, New Jersey
Leuzzi M.S.P., de Almeida F.S., Orsi M.L. and Sodre L.M.K., 2004, Analysis by RAPD of the genetic structure of Astyanax altiparanae (Pisces, Characiformes) in reservoirs on the Paranapanema River, Brazil, Genetics and Molecular Biology, 27: 355-362
http://dx.doi.org/10.1590/S1415-47572004000300009
Lewontin R.C., 1972, The apportionment of human diversity, Evolutionary Biology, 6:381-398
http://dx.doi.org/10.1007/978-1-4684-9063-3_14
Magurran A.E., 2004,Measuring biological diversity, Blackwell Publishing, USA
McDonald B.A. and McDermott J.M., 1993, Gene flow in plant pathosystems, Annual Review of Phytopathology, 31:353-73
http://dx.doi.org/10.1146/annurev.py.31.090193.002033
Meffe G.K. and Carroll G.R., 1997, Principles of Conservation Biology, Sinauer Associates, Sunderland, MA
Miller M.P., 1997, Tools for population genetic analysis (TFPGA) 1.3: A windows program for the analysis of allozyme and molecular population genetic data
Mukhopadhyay T. and Bhattacharjee S, 2014a, Study of the Genetic diversity of the ornamental fish Badis badis (Hamilton-Buchanan, 1822) in the Terai region of sub-Himalayan West Bengal, India, International Journal of Biodivesity, doi: 10.1155/2014/791364
http://dx.doi.org/10.1155/2014/791364
Mukhopadhyay T. and Bhattacharjee S., 2014b, Standardization of genomic DNA isolation from minute quantities of fish scales and fins amenable to RAPD-PCR, Proceedings of Zoological Society, 67 (1): 28-32
http://dx.doi.org/10.1007/s12595-013-0065-4
Nei M., 1978, Estimation of average heterozygosity and genetic distance from a small number of individuals, Genetics, 89:583-590
Peakall R. and Smouse P.E., 2012, GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update, Bioinformatics 28: 2537-2539
http://dx.doi.org/10.1093/bioinformatics/bts460
Peakall R. and Smouse P.E., 2006, GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research, Molecular Ecology Notes, 6: 288-295
http://dx.doi.org/10.1111/j.1471-8286.2005.01155.x
Prior K.A., Gibbs H.L. and Weatherhead P.J., 1997, Population Genetic Structure in the Black Rat Snake: Implications for Management, Conservation Biology, 1147-1158
http://dx.doi.org/10.1046/j.1523-1739.1997.96098.x
Rambaut A. and Drummond A., 2010, FigTree v1.3.1. [http://tree. bio.ed.ac.uk/software/figtree/]. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, United Kingdom
Talwar P.K. and Jhingran A.G., 1991, Inland Fishes of India and Adjacent countries, Oxford and IBH Co. Pvt. Ltd. (New Delhi), India
Wasko A.P. and Galetti Jr. P.M., 2002, RAPD analysis in the Neotropical fish Brycon lundii: genetic diversity and its implications for the conservation of the species, Hydrobiologia, 474: 131-137
http://dx.doi.org/10.1023/A:1016569919615
Wright S., 1969, Evolution and the Genetics of Populations, University of Chicago Press, Chicago
Yamazaki Y., Fukutomi N., Oda N., Shibukawa A., Niimuran Y. and Iwata A., 2005, Occurrence of larval Pacific lamprey Entosphenus tridentatus from Japan, detected by random amplified polymorphic DNA (RAPD) analysis, Ichthyological Research, 52: 297-301
http://dx.doi.org/10.1007/s10228-005-0276-4
Yeh F.C., Yang R.C., Boyle T.B.J., Ye Z.H. and Mao J. X., 1999, POPGENE version 1.32, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada (http://www.ualberta.ca/fyeh/)
. PDF(1089KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Tanmay Mukhopadhyay
. Soumen Bhattacharjee
Related articles
. Badis badis
. Threatened ornamental fish
. SHE analysis
. Genetic hierarchy
. RAPD fingerprinting
Tools
. Email to a friend
. Post a comment
.jpg)
.jpg)

.png)
.png)


